Based on the polarity of double stranded DNA molecule and characteristic of DNA polymerase III Holoenzyme that can only synthesize DNA in 5′-3′ direction, replication is carried in a discontinuous manner. There are two core polymerase on the replisome and each core takes part in leading or lagging synthesis. On the leading template, the new strand is synthesized continuously in the same direction with the fork progression, while on lagging template, the synthesis occurs in the opposite direction with fork movement. Therefore, the synthesis of lagging strand is discontinuous, requires many RNA primers and the recycling of lagging strand polymerase, resulting a number of Okazaki fragment (each Okazaki fragment requires one RNA primer). Basically, the molecular basis of DNA replication have been widely studied. However, the mechanisms of lagging strand synthesis is not clear yet and still controversial. For example, what is the trigger of lagging strand polymerase recycling, or how the next Okazaki fragment is synthesized?
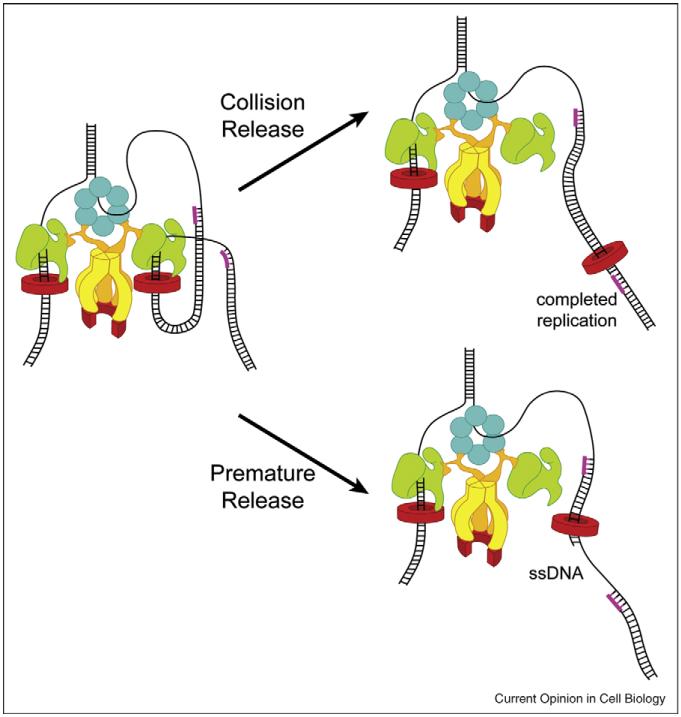
In the first model, named as “collision release” model, when the lagging polymerase collides the end of the previous Okazaki fragment, it will trigger dissociation of the polymerase from the clamp, leaving the clamp alone on DNA. After that, this free polymerase will bind to a new clamp, assembled on a new RNA primer by the clamp loader, to start synthesis of the next Okazaki fragment. In this model, there will be no gap between two Okazaki fragments.
Reference:
Jingsong Yang et al. The Control Mechanism for Lagging Strand Polymerase Recycling during Bacteriophage T4 DNA Replication. Molecular Cell, 2006.