Replisome is a multiple protein complex that works together on the template in order to synthesize new DNA. Basically, E. coli replisome contains a hexamer helicase, single stranded binding protein SSB, primase DnaG, 2 Polymerase III core, a gamma complex (also known as clamp loader) and β clamp.
Helicase is a homohexamer that encircles the lagging strand like a ring. It translocates along the ssDNA and separate the duplex DNA using energy from ATP hydrolysis. The unwinding of double stranded DNA and discontinuous synthesis of lagging strand leaves a ssDNA bubble behind the replication fork, on lagging template. This ssDNA is coated by SSB. Primase DnaG transiently binds to helicase to synthesize short RNA primers those are required for each Okazaki fragment synthesis of lagging strand, and dissociates when one primer synthesis is finished.
There are two DNA Polymerase III called twin Pol III core, each takes part in replication of leading and lagging strand, known as leading Pol III core and lagging Pol III core. Each core consists of 3 subunits: α, ε and θ. These two cores attach to a clamp loader δδ’2τγψχ complex by the τ subunit. This enables to synthesis of leading and lagging strands at the same time. The τ subunit of Pol III core also interacts with helicase. The assembly contains a clamp loader δδ’2τγψχ and two Pol III core is called as Pol III*.
Each core of Pol III* binds to a β clamp through the α subunit. β clamp is loaded onto ssDNA by the clamp loader δδ’2τγψχ. Because β clamp also has a ring shape that can translocate along each singled stranded leading and lagging template, the binding of Pol III core to the clamp keeps Pol III core stays on DNA for longer time. That is why Pol III can synthesize with very high processivity (more than 5 kb per binding event). Pol III* carrying β clamp is referred as Pol III holoenzyme.
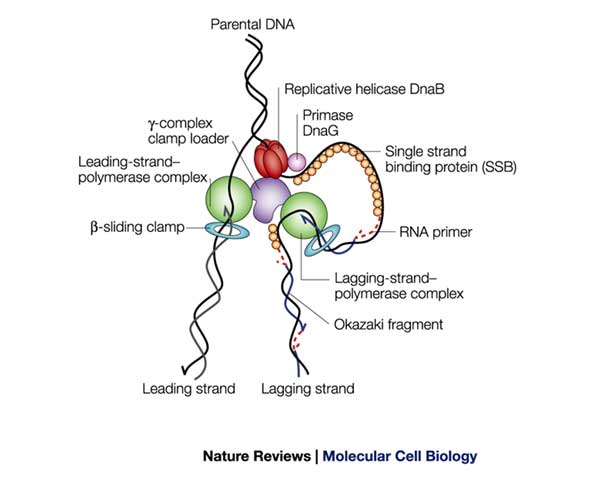
Replisome structure. (Photo referred from Nature Reviews Molecular Cell Biology 3, 859-870, November 2002, doi:10.1038/nrm951)